Note
Go to the end to download the full example code
5.4. Coverage module example
from sequana import SequanaCoverage, sequana_data
bedfile = sequana_data("JB409847.bed")
Reading input BED file
gc = SequanaCoverage(bedfile)
0it [00:00, ?it/s]
1it [00:00, 111.46it/s]
Select a chromosome (first and only one in that example)
chrom = gc[0]
print(chrom)
Chromosome length: 19795
Sequencing depth (DOC): 931.31
Sequencing depth (median): 988.00
Breadth of coverage (BOC) (percent): 96.60
Coverage standard deviation: 237.15
Coverage coefficient variation: 0.25
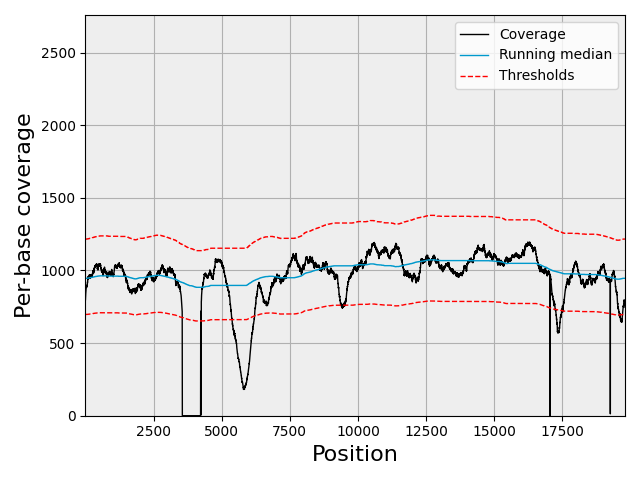
Compute running median and zscore telling the algorithm that the chromosome is circular.
chrom.running_median(n=5001, circular=True)
chrom.compute_zscore()
print(chrom.get_centralness())
0.8684516291992928
Plotting
chrom.plot_coverage()
Total running time of the script: (0 minutes 0.801 seconds)