4.5.6. quality_control
current version:?
This pipeline is not maintained anymore but should be functional. It is a short-read quality control pipeline from the Sequana project. We would recommend to use the fastqc, demultiplex, and multitax pipelines instead.
| Overview: | A quality control pipeline for illumina data set. This pipeline removes contaminants (e.g. Phix), performs fastqc, adapter cleaning and trimming and checks for contaminants |
|---|---|
| Input: | Raw fastq files |
| Output: | Cleaned fastQ files, remove phix and adapters + taxonomy |
| Status: | production. not maintained. Please use sequana_fastqc and sequana_multitax pipeline instead |
| Citation: | Cokelaer et al, (2017), ‘Sequana’: a Set of Snakemake NGS pipelines, Journal of Open Source Software, 2(16), 352, JOSS DOI doi:10.21105/joss.00352 |
Installation
You must install Sequana first:
pip install sequana
Then, just install this package:
pip install sequana_quality_control
Usage
sequana_quality_control --help sequana_quality_control --input-directory DATAPATH
This creates a directory with the pipeline and configuration file. You will then need to execute the pipeline:
cd quality_control sh quality_control.sh # for a local run
This launch a snakemake pipeline. If you are familiar with snakemake, you can retrieve the pipeline itself and its configuration files and then execute the pipeline yourself with specific parameters:
snakemake -s quality_control.rules -c config.yaml --cores 4 --stats stats.txt
Or use sequanix interface.
Requirements
This pipelines requires the following executable(s):
- fastqc
- bwa
- sambamba
- samtools
- pigz
- cutadapt [or atropos]
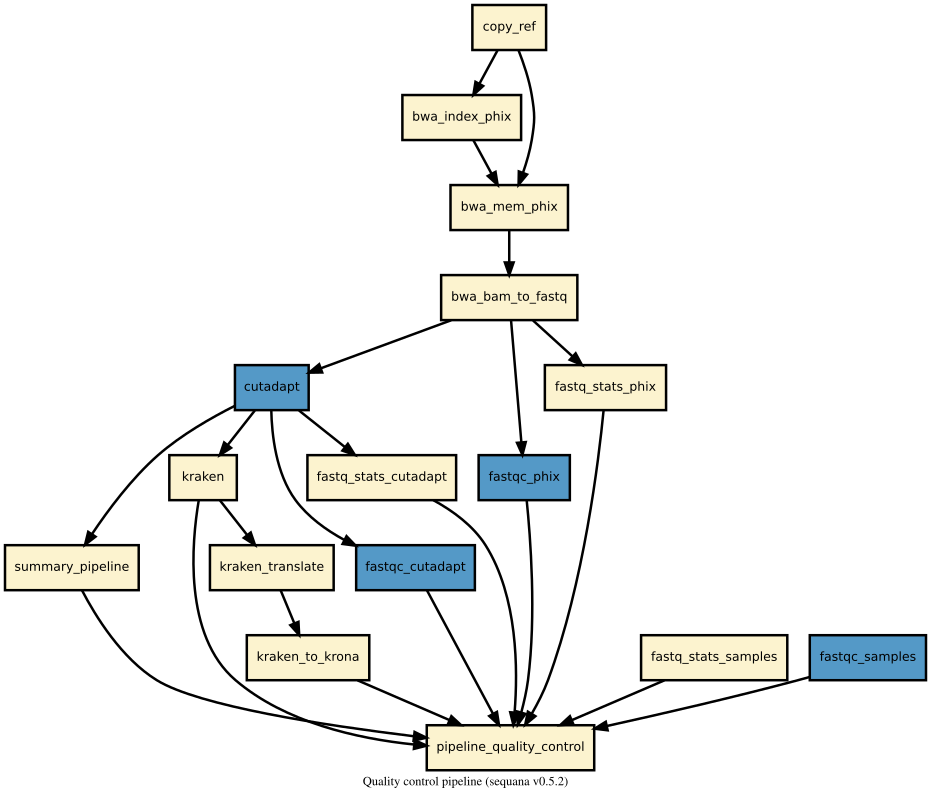
Details
This pipeline runs quality_control in parallel on the input fastq files (paired or not). A brief sequana summary report is also produced.
Rules and configuration details
Here is the latest documented configuration file to be used with the pipeline. Each rule used in the pipeline may have a section in the configuration file.
ChangeLog
| Version | Description |
|---|---|
| 0.11.0 |
|
| 0.10.0 |
|
| 0.9.0 |
|
| 0.8.4 |
|
| 0.8.3 |
|
| 0.8.2 |
|
| 0.8.1 | uses more sequana tools to handle the options |